Molecular Vectorology
AAV vectors are tools of choice for in vivo gene transfer. Their efficiency is well established with the success of numerous clinical trials and the commercialization of two products, Luxturna® and Zolgensma®. However, the injection of high doses of viral vectors for the treatment of systemic diseases poses new challenges that include the tissue specificity and the immune response directed against the vectors. Indeed, an immune response can appear against the viral capsid, the foreign DNA or the therapeutic protein. Impurities may also be involved in the detection of the product by the immune system.
Our team is developing analytical methods to characterize rAAV genomes by high-throughput sequencing (HTS) and studying the influence of DNA sequences on the efficiency of AAV viral vectors.
Characterization of nucleic acids by high-throughput sequencing
In our laboratory, we have developed a method based on high-throughput sequencing (HTS) and a dedicated bioinformatics pipeline to analyze the residual DNA and the identity of rAAV genomes. This method called SSV-Seq (Single-Stranded DNA Virus Sequencing) has been published (Lecomte et al. 2015, 2019) and is detailed on the page link to SSV-Seq page.
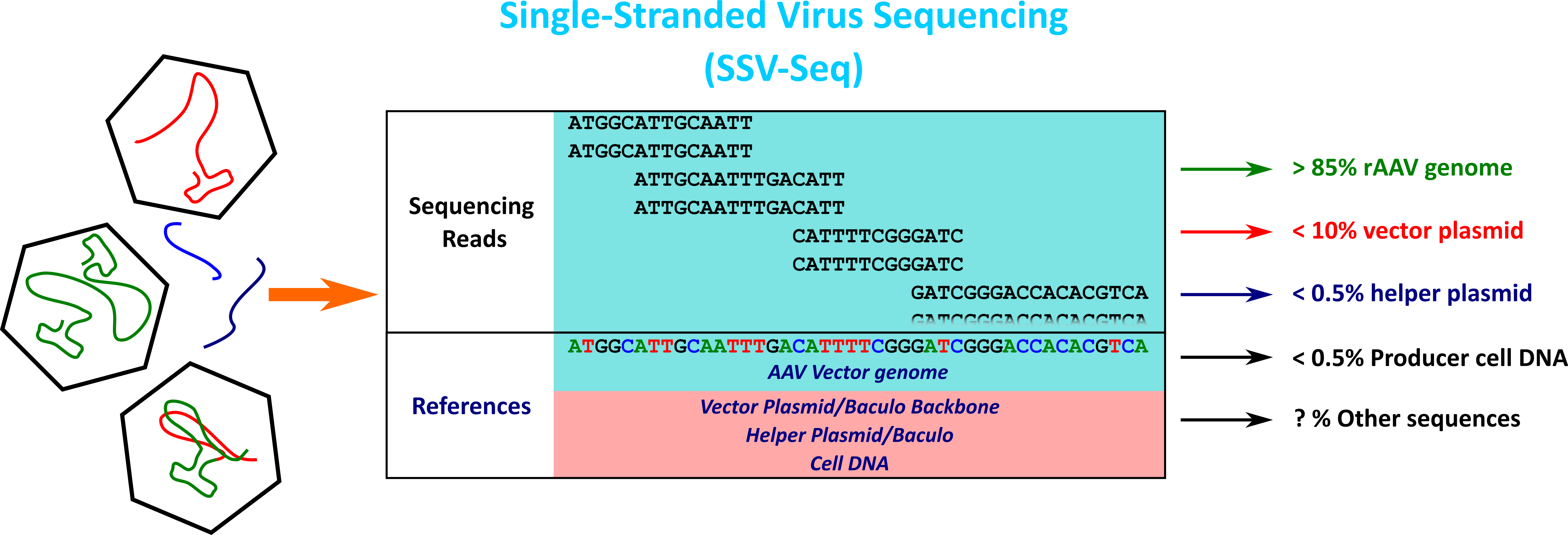
Contamination of the rAAV batches by residual RNA, in particular by cellular microRNAs, is also studied in order to avoid a potential risk for the patient.
Optimization of the rAAV vector genomes
The vector potency and specificity depend on the choice of the capsid variant and on the design of the rAAV genome (promoter, enhancers, etc.). Our research projects are particularly focused on the role of the terminal repeated sequences (ITR), the only viral sequences present in vectors, during gene transfer.
Team Members :
Magalie Penaud-Budloo, Ph.D., Senior scientistEmilie Lecomte, Bioinformatics scientist
Sylvie Saleun, Engineer
Marianne Laugel, PhD student